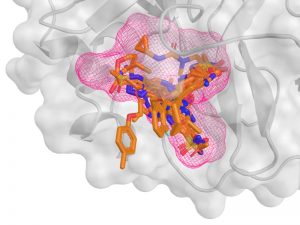
 Screening & affinities of potential inhibitors of SARS-CoV2 proteases. Kozakov group seek inhibitors of the viral proteases that are necessary for viral maturation. Starting with the hotspot map of the target generated by FTMAP, they use FTScreen for ultrafast virtual screen of extra-large (>109) commercially available combinatorial libraries of small organic molecules, followed by a more rigorous analysis of the best fits using the broad repertoire of computational methods, including our award-winning MCM and MELD × MD in collaboration with Dill group. The compounds are tested in vitro by Tonge group.
Screening & affinities of potential inhibitors of SARS-CoV2 proteases. Kozakov group seek inhibitors of the viral proteases that are necessary for viral maturation. Starting with the hotspot map of the target generated by FTMAP, they use FTScreen for ultrafast virtual screen of extra-large (>109) commercially available combinatorial libraries of small organic molecules, followed by a more rigorous analysis of the best fits using the broad repertoire of computational methods, including our award-winning MCM and MELD × MD in collaboration with Dill group. The compounds are tested in vitro by Tonge group.
 Designing a PROTAC linker to target viral proteins for degradation. PROteolysis-TArgeting Chimeras (PROTACs) exploit the intracellular ubiquitin-proteasome system to selectively degrade target proteins. Kozakov group focus their efforts on development of PROTACs whose structure and geometry is optimal for fast and efficient ubiquitination of SARS-CoV-2 proteases. In this study, they employ their award-winning protein docking software ClusPro and LigTBM fot ligand docking. The predictions are refined by the Dill group. Predicted PROTACs are tested experimentally by the Peter Tonge group at SBU.
Designing a PROTAC linker to target viral proteins for degradation. PROteolysis-TArgeting Chimeras (PROTACs) exploit the intracellular ubiquitin-proteasome system to selectively degrade target proteins. Kozakov group focus their efforts on development of PROTACs whose structure and geometry is optimal for fast and efficient ubiquitination of SARS-CoV-2 proteases. In this study, they employ their award-winning protein docking software ClusPro and LigTBM fot ligand docking. The predictions are refined by the Dill group. Predicted PROTACs are tested experimentally by the Peter Tonge group at SBU.
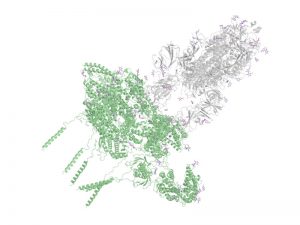 Modeling the structural interactome of SARS-CoV-2. The cell invasion and subsequent replication of SARS-CoV-2 involves a significant degree of interaction between the viral and human proteins. Cutting edge research provides experimental evidence for a number of such interactions. However, the atomistic structural detail of the protein-protein complexes mediating these interactions is, at this point, largely missing. We aim to bridge this gap and construct the models of various complexes formed by SARS-CoV-2 and human protein by utilizing the full extent of structure modeling tools developed in the lab.
Modeling the structural interactome of SARS-CoV-2. The cell invasion and subsequent replication of SARS-CoV-2 involves a significant degree of interaction between the viral and human proteins. Cutting edge research provides experimental evidence for a number of such interactions. However, the atomistic structural detail of the protein-protein complexes mediating these interactions is, at this point, largely missing. We aim to bridge this gap and construct the models of various complexes formed by SARS-CoV-2 and human protein by utilizing the full extent of structure modeling tools developed in the lab.