We present an ultra-fast approach to modeling protein interactions.
Protein-protein interactions (PPIs) are the basis of cellular functions, and when these processes are compromised diseases such as cancer emerge. For years scientists have tried with mixed success to map out PPIs to understand cellular processes. Now our group has outlined a method that could pave the way to designing new drugs that prevent problematic protein interactions that lead to disease. The findings are published in the early online edition of PNAS.
Proteins are the major building blocks of the cell. Many proteins perform their function by interacting with other proteins. In a typical cell, hundreds of thousands of different protein interactions take place. Characterizing the structure of these interactions helps elucidate how organisms function normally and during disease development.
The problem considered is given three dimensional structures of two individual proteins to predict how these protein interact with each other. We can liken the method to characterizing all the possible structures that pairs of “lego blocks” form out of a huge set of different individual starting blocks.
In the paper titled “Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds,” we explain a new algorithm used to create ultra-fast approach to modeling protein interactions. We discovered that the method runs an order of magnitude faster than previous state-of-the-art methods and has comparable accuracy.
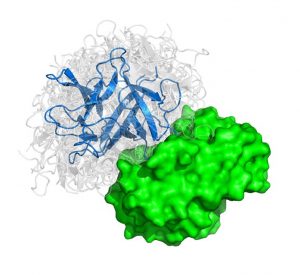
The algorithm features a fast Manifold Fourier transform (FMFT) that helps to speed the calculations, enabling us to sample a large number of putative protein-protein complex conformations.
The new algorithm will soon be available to the scientific community through our publicly available protein-protein docking server called ClusPro. This resource, with more than 15000 academic users worldwide, supported by the National Science Foundation and the Binational Science Foundation, is being developed by our group in collaboration with scientists at Boston University. ClusPro was judged to be the best automated docking server in the latest rounds of the international blind protein docking competition called CAPRI (Critical Assessment of Prediction Interaction).