Dr. Kozakov and Dr. Padhorny in collaboration with researchers from Boston University and Boston National Emerging Infectious Diseases Laboratories (NEIDL), have analyzed the difference in phosphorylation patterns between SARS-COV2 virus-infected and healthy alveolar lung cell (AT2). This survey revealed 4,688 differential phosphosites mapping to 1,166 unique proteins, which were clustered into distinct clusters based on temporal enrichment, associated with protein domains and cellular processes linked to infection, such as viral messenger RNA synthesis and export of viral ribonucleoproteins, as an immediate response to SARS- CoV-2 entry. Our group has performed in silico structural modeling of experimentally observed viral-host protein-protein interactions, using award-winning computational tools developed in our lab. That enabled us to structurally characterize the interactions that were detected in MS experiments and independently corroborate experimental observations. Our modeling identified several key types of proteins that dominated these interactions, including the kinases of GSK3, MAPK, and CK1 families, and a number of other targets. We hypothesized that modulating those targets might have antiviral effect. We have identified and modeled interactions of several clinically safe compounds from BROAD database targeting those families using our award wining LigTBM molecular modeling approach. Six of the selected compounds have efficiently inhibited viral replication by more than 90% in the AT2 lung cells. The paper has appeared in Molecular Cell.
Author: Andrey Alekseenko
COVID-19 efforts
During the current crisis, it is everyone’s responsibility to make their best efforts. Our group is now targeting the research towards the search of new compounds targeting SARS-CoV-2 proteins. We tightly collaborate with experimental and computational groups on these projects. Besides doing our part in helping with the current epidemic, we hope that the methods and tools developed will allow faster response to future viral threats. More information is available on a dedicated page: https://abcgroup.cluspro.org/research/covid-19/
Our team has been awarded IACS SEED grant
On April 28th, the results of the IACS Seed Grant competition were announced. The joint project of our group and Dr. Rezaul Chowdhury, “Speeding up flexible peptide-protein docking using convolutional neural networks,” was chosen as one of the two winners. We’re grateful for this support and are happy that the grant committee shares our belief in the idea of applying state-of-the-art deep learning methodology to the notorious for its complexity important scientific and medical problem of structure prediction of protein-peptide complexes.
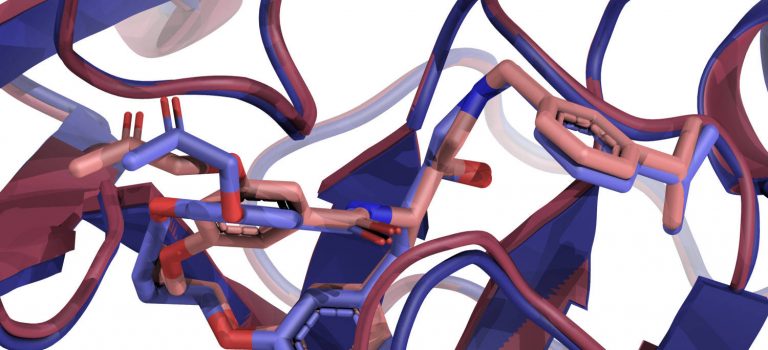
Our LigTBM ligand docking approach is top performer in D3R Grand Challenge 4 Blind docking competition
D3R (Drug Design Data Resource) Grand Challenge is a blinded prediction challenge for the computational chemistry community, with components addressing pose-prediction, affinity ranking, and free energy calculations. Its fourth installment, D3R GC4, was held from October to December 2018.
Our group participated in the pose prediction challenge for the macrocyclic inhibitors for Beta secretase 1 (BACE1). This protein is involved in the generation of beta-amyloid peptides and presents an important target for developing drugs for Alzheimer’s disease. In the stage 1a of the challenge, the organizers presented participants with the apo-structure of the receptor and SMILES strings describing 20 ligands.
According to the official rankings, the template-based method developed in our group scored best, out of 74 total submitted entries, in terms of the Mean RMSD. Our group achieved sub-angstrom mean and median RMSD for this challenge.
Our method relies on finding structures of distant homologs of the target protein that bind similar ligands, and using them at all stages of the protocol: initial pose generation, structure refinement, and the final scoring. More details are available in our paper, published in the Journal of Computer-Aided Molecular Design.
We refined and automated the approach used in this challenge. It is now available for free academic and non-commercial use as a user-friendly web-server, ClusPro LigTBM. This version of the protocol is described in our Journal of Molecular Biology paper.